DNA-extraction



Based on our experience, we select the optimum variant for the respective sample type from various extraction methods in order to obtain high-quality DNA (and RNA on request).
All DNA extractions are carried out automatically to rule out contamination and guarantee the highest quality.
Diagnostic-PCR
With diagnostic PCR, even the smallest amounts of target DNA can be detected. This allows the presence or absence of a species to be checked quickly and accurately. Diagnostic PCR is based on the use of specific probes that exactly match the DNA being searched for. If the target organism is present, the sample is positive; if it is absent, it is negative. All of the diagnostic approaches we use are optimized in terms of their specificity and sensitivity.
Sinsoma offers three different PCR methods for the diagnostic detection of species, enabling us to offer our customers the optimum solution for their specific problem:
- qPCR (Quantitative PCR)
- cePCR (Endpoint PCR with capillary electrophoresis)
- ddPCR (Droplet Digital PCR)
For a targeted search for species


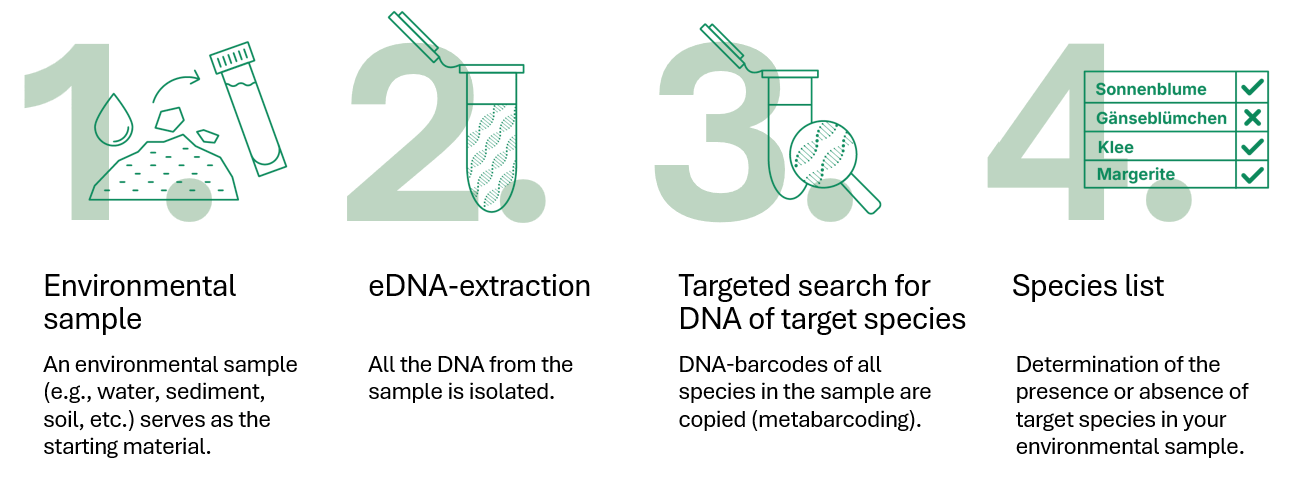
- An environmental sample (e.g. water, sediment, soil, etc.) serves as the starting material.
- The entire eDNA of the sample is extracted.
- Search for DNA of one or more target species using specific and highly sensitive molecular probes.
- Determination of the presence or absence of the target species(es) in your environmental samples.
The result
- Report including an overview of the methodological approach used.
- Information on the presence/absence of the target species you are looking for in your samples
- Excel table, which provides the data basis for measures to protect health and the environment.
Diagnostic PCR is suitable for the detection of
- Invasive species (e.g. signal crayfish, quagga mussel)
- Protected species, FFH species (e.g. mudminnow, moor frog, crested newt)
- Pathogens (e.g. crayfish plague, amphibian fungi Bd, Bsal)
- Parasites (e.g. bedbug, varroa mite)
DNA-barcoding
Similar to recognizing products by scanning the barcode in the supermarket, the identification of a species is based on selected DNA sections. These so-called DNA barcodes allow clear and reliable species identification of animals, plants, fungi and bacteria. Molecular species identification is independent of the developmental stage (egg, larva, adult).
For DNA-based species identification



- The eDNA of an unknown organism is extracted.
- A specific DNA section that is well suited for identification is amplified and read.
- Bioinformatic analysis to identify the organism by comparison with sequence databases.
Your result
- Report, incl. overview of the methodological approach used.
- List of identified species as an Excel table.
- A DNA-based species certificate on request.
DNA-METABARCODING
In DNA metabarcoding, the entire diversity of species in a sample is analyzed and evaluated at once. This method is used for samples that contain DNA from several species (e.g. water, honey, soil, dust, feces). It allows the simultaneous determination of different animal, plant, bacterial or fungal species from environmental or food samples.
DNA metabarcoding makes biodiversity measurable. This makes it a powerful tool, both for biodiversity studies and for achieving a company’s SDGs. This method can be used to identify the composition of species communities that occur in a particular habitat. Repeated sampling can also be used to record changes over time or evaluate an actual state before and after measures have been taken.
For the survey of the species spectrum


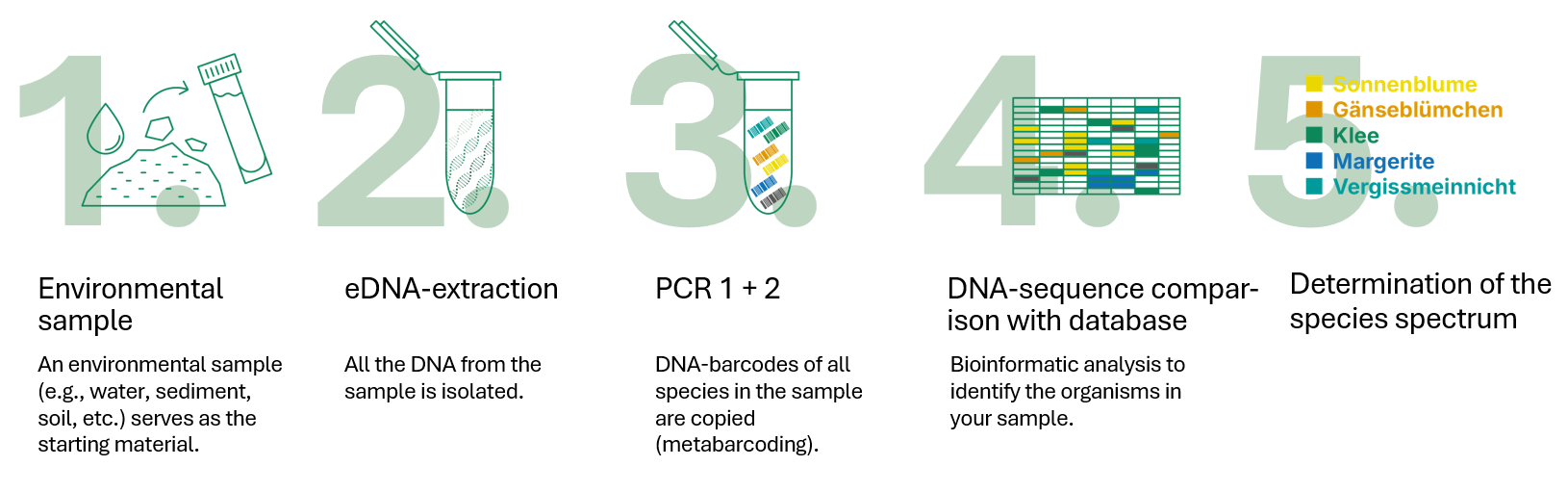
- An environmental sample (e.g. water sample, sediment, soil) serves as the starting material.
- The entire DNA of the sample is extracted.
- Search for the DNA of one or more groups of organisms (e.g. fish, plants) using metabarcoding.
- Bioinformatic analysis to identify the organisms in your environmental samples.
- Determination of the presence of a specific target species in your environmental sample.
Your result
- Comprehensible report that provides the data basis for measures to protect health and the environment.
- Overview of the methodological approach used.
- Information on the species spectrum found in your sample, including the sequence number.
- Interactive pie charts for graphical exploration of the species composition (Krona Charts).
Metabarcoding is suitable for
- To make biodiversity measurable
- For biodiversity monitoring and nature conservation
- For SDG reporting
- To review renaturation and conservation measures
- To identify food spectra